Datasets
Find below some published datasets that I was involved.
Gas sensors for home activity monitoring Data Set
Available on UCI Machine Learning Repository
Authors: Flavia Huerta, Ramon Huerta, Thiago Mosqueiro, Jordi Fonollosa, Nikolai Rulkov, Irene Rodriguez-Lujan
This dataset has recordings of a gas sensor array composed of 8 MOX gas sensors, and a temperature and humidity sensor. This sensor array was exposed to background home activity while subject to two different stimuli: wine and banana. The responses to banana and wine stimuli were recorded by placing the stimulus close to the sensors.
Please cite: Online decorrelation of humidity and temperature in chemical sensors for continuous monitoring. Chemometrics and Intelligent Laboratory Systems 2016.
Electrical pulses of two electric fish swimming freely in the same aquarium
Authors: R. F. Guariento, T. Mosqueiro, P. Matias, V. B. Cesarino, L. O. B. Almeida, J. F. W. Slaets, L. P. Maia, R. D. Pinto.
In this dataset, we are sharing a time series of of electric pulses from dyads of freely moving Gymnotus sp. Because both fish are completely free to swim, the challenge is to detect which pulses were discharged by each of the mfish. A rich repertoire of electrical behaviors can be observed, such as (i) offs (moments of silence) and (ii) chirps (small and noisy oscillations). For more information, visit our github.
Please cite:
- Automated pulse discrimination of two freely-swimming weakly electric fish and analysis of their electrical behavior during a dominance contes. Under review.
- Non-parametric Change Point Detection for Spike Trains. Accepted talk on CISS (Princeton 2016). Published on IEEE
Task allocation and site fidelity synergistically influence foraging regulation in honey bee colonies
Authors: Chelsea Cook, Thiago Mosqueiro, Ramon Huerta, Jurgen Gadau, Brian Smith, Noa Pinter-Wollman.
In this dataset we are sharing behavioral observations of honey bees foraging for two feeders placed near their hive. This data was used to assess the persistence of honey bees at foraging for a particular resource location when presented with two different feeders with sucrose solution.
Please cite: Mosqueiro T, Cook C, Huerta R, Gadau J, Smith B, Pinter-Wollman N. 2017 Task allocation and site fidelity jointly influence foraging regulation in honeybee colonies. R. Soc. open sci. 4:170344. DOI: 10.1098/rsos.170344 (in press). Preprint Here.
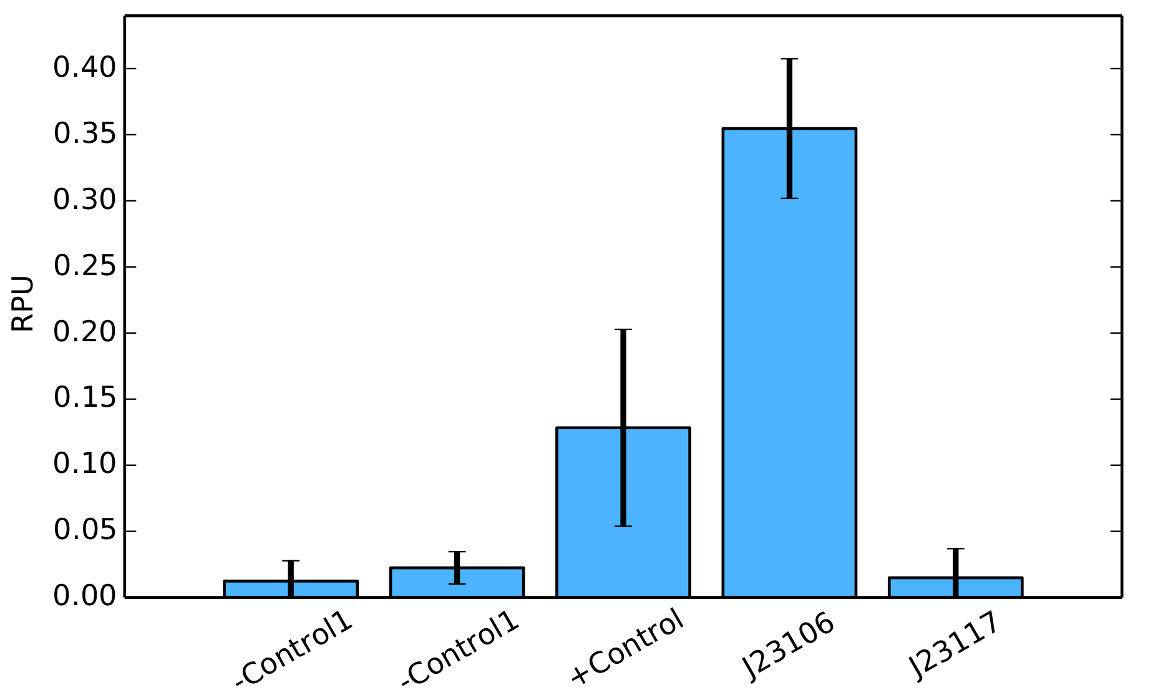
iGEM 2015 Interlab Experiment
The Second International InterLab Measurement Study in synthetic biology proposes to detect three promoters activity by measuring and comparing the levels of GFP expression of three devices expressing GFP and controlled by one of the three promoters (see Figure 1). The three promoters - BBa_J23101, BBa_J23106, BBa_J23117 - are from the Anderson library, a constitutive promoter family with different strengths.
Please cite:
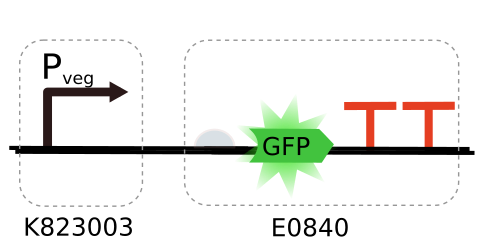
iGEM 2015 pVeg promoter in E. Coli
Pveg is a strong constitutive promoter well studied in Bacillus subtilis, especially during the vegetative growth phase and sporulation (controlled primarily by the veg gene). Currently available at Registry as BioBrick K823003, it was characterized using a β-galactosidase assay by team LMU-Munich, as well as published in a recent paper [1]. While originally designed for B. Subtilis, during the 2014 iGEM edition Brasil-SP team proposed a protocol to express it in E. Coli (DH5α strain). Repeating this procedure with a gfp gene, we can now perform experiments similar to the β-galactosidase assays and evaluate how strong Pveg is in E. Coli.
Please cite: